To sequence a genome for the first time is still not a simple endeavour.
Maybe that is why, out of more than 1.5 million species described, we have less than 1000 genomes sequenced in the genbank database.
So, the odds are, if you are interested in the genome of an organism to know more about its behaviour, physiology, ecology and life history, you will have to sequence it yourself.
And that was the case for our species of interest, the sun coral Tubastrea sp.
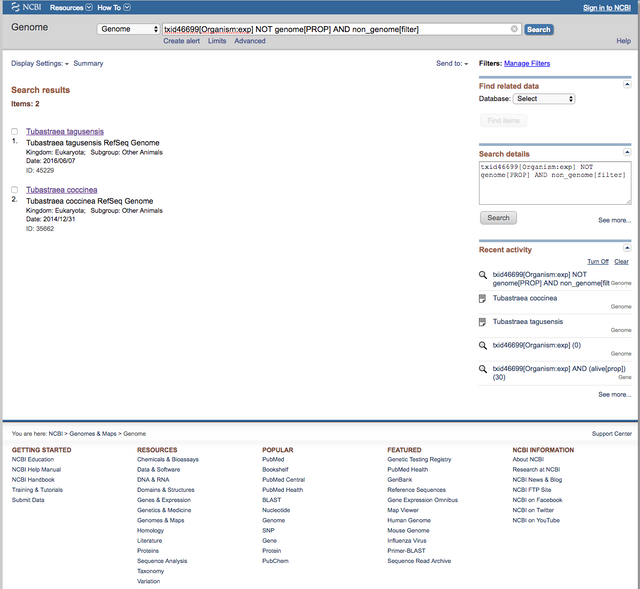
Screeshots from genbank shows that we have sequenced only 103 genes sequenced for this genus: 39 for the main species, T. coccinea, 17 for T. micranthus and 10 for T. aurea.
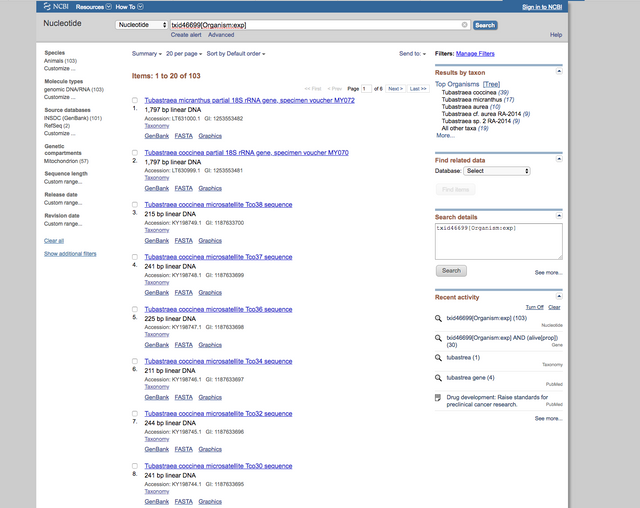
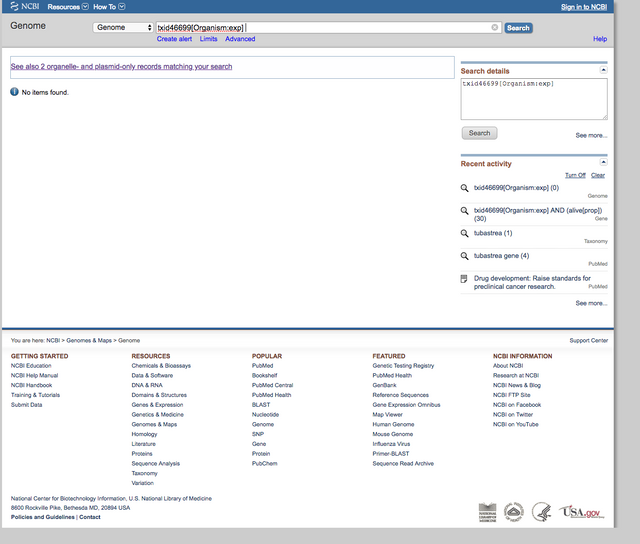
It was even surprising to find the mitochondrial genomes of the 2 main species: T. coccinea and T. micranthus
Capel KCC., Migotto AE., Zilberberg C., Lin MF., Forsman Z., Miller DJ., Kitahara M V. 2016. Complete mitochondrial genome sequences of Atlantic representatives of the invasive Pacific coral species Tubastraea coccinea and T. tagusensis (Scleractinia, Dendrophylliidae): Implications for species identification. Gene 590:270–277. DOI: 10.1016/j.gene.2016.05.034.
Even though the price of sequencing a human genome is dropping, the price of sequencing an animal genome for the first time is still high. The estimated cost of the Pacific Oyster genome, published in 2012, was USD 2 million.
Wang J., Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., Yang P., Zhang L., Wang X., Qi H., Xiong Z., Que H., Xie Y., Holland PWH., Paps J., Zhu Y., Wu F., Chen Y., Wang J., Peng C., Meng J., Yang L., Liu J., Wen B., Zhang N., Huang Z., Zhu Q., Feng Y., Mount A., Hedgecock D., Xu Z., Liu Y., Domazet-Lošo T., Du Y., Sun X., Zhang S., Liu B., Cheng P., Jiang X., Li J., Fan D., Wang W., Fu W., Wang T., Wang B., Zhang J., Peng Z., Li Y., Li N., Wang J., Chen M., He Y., Tan F., Song X., Zheng Q., Huang R., Yang H., Du X., Chen L., Yang M., Gaffney PM., Wang S., Luo L., She Z., Ming Y., Huang W., Zhang S., Huang B., Zhang Y., Qu T., Ni P., Miao G., Wang J., Wang Q., Steinberg CEW., Wang H., Li N., Qian L., Zhang G., Li Y., Yang H., Liu X., Yin Y., Wang J. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. DOI: 10.1038/nature11413.
We never calculated exactly the cost of sequencing the genome of the Golden Mussel Limnoperna fortunei. However, I estimated the number of work hours dedicated by the researchers involved in the project.
It consumed 42,000 hours, by 12 direct researchers from 7 institutions over the course of 8 years.
Uliano-Silva M., Americo JA., Brindeiro R., Dondero F., Prosdocimi F., Rebelo MDF. 2014. Gene discovery through transcriptome sequencing for the invasive mussel Limnoperna fortunei. PLoS ONE 9. DOI: 10.1371/journal.pone.0102973.
Uliano-Silva M., Americo JA., Costa I., Schomaker-Bastos A., de Freitas Rebelo M., Prosdocimi F. 2016. The complete mitochondrial genome of the golden mussel Limnoperna fortunei and comparative mitogenomics of Mytilidae. Gene 577:202–208. DOI: 10.1016/j.gene.2015.11.043.
Uliano-Silva M., Dondero F., Dan Otto T., Costa I., Lima NCB., Americo JA., Mazzoni CJ., Prosdocimi F., Rebelo M de F. 2018. A hybrid-hierarchical genome assembly strategy to sequence the invasive golden mussel, Limnoperna fortunei. GigaScience 7:1–10. DOI: 10.1093/gigascience/gix128.
Yes, it could have probably been less money and hours today, if we didn’t had to do it step by step. But that is what it took.
At the same time, it could take more. Some genomes are simply too hard to assemble and remain unassembled even after many years.
The benchmark work in other corals and cnidarians may help the accomplishment of the challengin task of assembling the sun coral genome.
Bhattacharya D., Agrawal S., Aranda M., Baumgarten S., Belcaid M., Drake JL., Erwin D., Foret S., Gates RD., Gruber DF., Kamel B., Lesser MP., Levy O., Liew YJ., MacManes M., Mass T., Medina M., Mehr S., Meyer E., Price DC., Putnam HM., Qiu H., Shinzato C., Shoguchi E., Stokes AJ., Tambutt� S., Tchernov D., Voolstra CR., Wagner N., Walker CW., Weber APM., Weis V., Zelzion E., Zoccola D., Falkowski PG. 2016. Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5:1–26. DOI: 10.7554/eLife.13288.
Baumgarten S., Simakov O., Esherick LY., Liew YJ., Lehnert EM., Michell CT., Li Y., Hambleton EA., Guse A., Oates ME., Gough J., Weis VM., Aranda M., Pringle JR., Voolstra CR. 2015. The genome of Aiptasia , a sea anemone model for coral symbiosis. Proceedings of the National Academy of Sciences 112:11893–11898. DOI: 10.1073/pnas.1513318112.
Wang X., Liew YJ., Li Y., Zoccola D., Tambutte S., Aranda M. 2017. Draft genomes of the corallimorpharians Amplexidiscus fenestrafer and Discosoma sp. Molecular Ecology Resources 17:e187–e195. DOI: 10.1111/1755-0998.12680.
Devlin-Durante MK., Baums IB. 2017. Genome-wide survey of single-nucleotide polymorphisms reveals fine-scale population structure and signs of selection in the threatened Caribbean elkhorn coral, Acropora palmata. PeerJ 5:e4077. DOI: 10.7717/peerj.4077.
Bird C., Karl S., Mouse P., Toonen R. 2011. Detecting and measuring genetic differentiation. In: 31–55. DOI: 10.1201/b11113-4.


